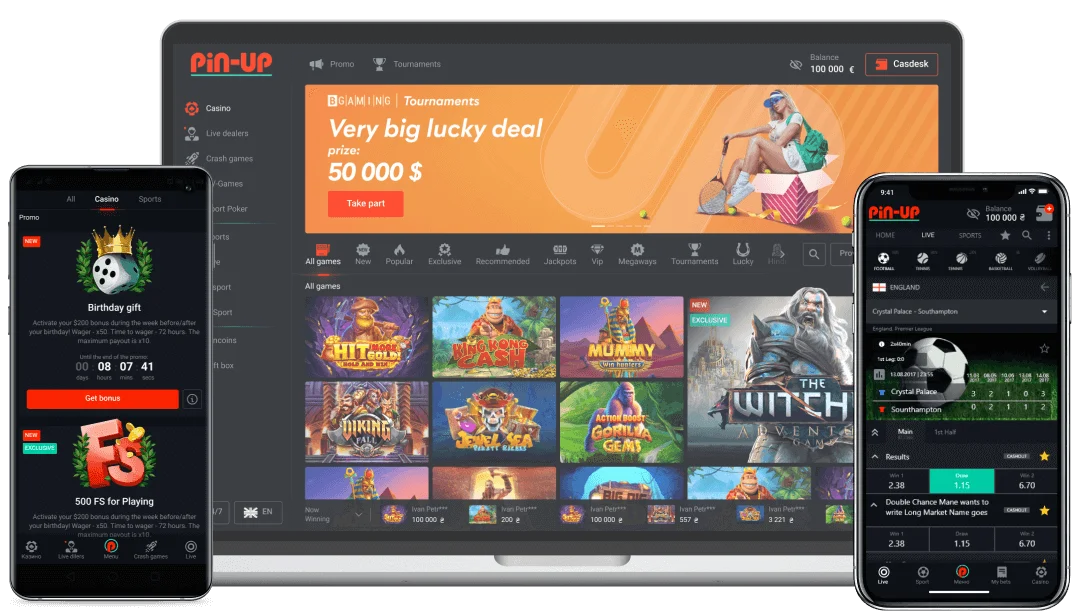
Pin-Up Online Entertainment in India

Live Casino

Fast Games

VIRTUAL SPORTS

ESPORTS
Table of Content
The digital entertainment market in India is rapidly evolving, creating space for modern gaming platforms that deliver a seamless user experience. Pin-Up stands out with its retro-inspired design, fast navigation, and diverse game catalogue tailored to both beginners and experienced players. As more Indian users migrate to online entertainment, the platform’s balance of speed, reliability, and mobile optimization becomes especially valuable. Before exploring categories such as Pin Up casino, users can review a concise information table that highlights key features, helping newcomers understand the fundamentals of the platform and prepare for a deeper dive into games, the app experience, and additional services.
|
Feature |
Description |
|
Brand |
Pin-Up |
|
Target Region |
India |
|
Main Category |
Online casino & mobile gaming |
|
Popular Game |
Aviator |
|
Popular Slots |
1000+ |
|
Mobile App |
Android & iOS |
|
Bonuses |
Casino, welcome, weekly, cashback |
|
Payment Methods |
UPI, PayTM, GPay, Cards |
|
Currency |
INR |
|
Audience |
18+ players |
|
Support |
24/7 |
|
Platform Type |
Licensed online operator |
|
Style |
Retro Pin-Up aesthetic |
|
Special Offers |
Pin Up bonus casino programs |
This table introduces key facts but is only the starting point. Below, the article explores features, game mechanics, mobile experience, bonuses and important insights that matter to Indian players today.
Pin Up Casino Overview for Indian Users
The online entertainment landscape in India is developing rapidly, and players increasingly expect platforms to deliver both convenience and depth. A growing number of users explore modern gaming environments that provide intuitive navigation, strong visual appeal, and stable performance across devices, which is where Pin Up Casino maintains a notable advantage. Instead of offering a generic set of features, the platform emphasizes personalized settings, smooth transactions in INR, and immediate switching between desktop, mobile browser, and app formats. Indian players consistently highlight the variety of available titles, the high RTP percentages, fast withdrawal processing, and the reliability of multilingual customer support.
For many newcomers, the best starting point is exploring the game library, especially because the system arranges content in a way that simplifies discovery for all experience levels. Pin-Up’s layered structure allows users to filter games by provider, volatility, mechanics, or popularity, making it easier to find preferred genres without unnecessary complexity. This approach is particularly valuable for players who wish to browse quickly while still maintaining control over their experience. The platform includes several core elements that contribute to its strong reputation, and the following breakdown helps clarify what users can expect when navigating these features.
Game Categories Available at Pin Up Casino
Pin-Up provides a diverse entertainment ecosystem, where players can try classic slot machines, live dealer tables, arcade-style crash games, and modern interactive experiences. Each category serves a different mood and gaming style, making the platform suitable for beginners and experienced players alike.
Before diving deeper, here are the most popular categories:
- Slots — the largest and most dynamic category.
- Live casino games — real-time tables with professional dealers.
- Crash games — including the widely famous Pin Up Casino Aviator.
- Table games — roulette, blackjack, baccarat and more.
- Instant games — quick picks with fast outcomes.
After selecting the preferred category, players can further personalize their experience through filters such as volatility, provider, and minimum bet size.
Pin-Up enriches each section with detailed statistics, return-to-player values, and demo modes, helping users develop confidence before placing real-money bets.
Pin Up App Mobile Experience and Benefits
The mobile segment of the gaming market in India continues to dominate, driven by the rapid adoption of smartphones and affordable internet access. Many users now expect platforms to provide smooth, stable performance on handheld devices, which has significantly shaped product development across the industry. Within this landscape, one of the strongest advantages is delivered through the Pin Up App, offering a mobile experience that enhances both speed and usability. More than 65% of Indian gamers prefer playing on their phones, and this preference is reflected in faster loading times, improved visual quality, and increased privacy for users who want seamless entertainment on the go.
Frequent travelers and those who enjoy short gaming sessions appreciate how the mobile-first interface adapts to different screen sizes and conditions without sacrificing performance. The platform also integrates exclusive promotional offers tailored specifically for app users, providing additional value that is not always available in the desktop version. Indian players benefit from a straightforward installation process and automatic updates, ensuring that features remain current, secure, and uninterrupted. This combination of convenience, personalization, and technical optimization is what strengthens the overall mobile identity of the platform and enhances long-term engagement.
Key Features of the Pin Up App
The mobile application is designed to meet the expectations of Indian users who value speed, convenience, and uninterrupted access to entertainment. Many modern platforms adapt to mobile trends, but only a few ensure full compatibility with varied devices and network conditions. This is where the Pin Up App demonstrates its advantage, offering flexible functionality that supports both casual and frequent gameplay.
Below are the most significant features that define the mobile experience:
- Intuitive interface optimized for small screens.
- Access to the full game catalogue, including Pin Up slot titles.
- Instant notifications for bonuses and tournaments.
- Secure digital wallet with UPI and PayTM support.
- High-speed loading for live casino streams.
These elements collectively create a seamless user journey, allowing players to enjoy stable performance even on older smartphones or slower internet connections. The balanced technical foundation and user-centric design ensure that entertainment remains accessible and smooth at any time. In a recent survey of online gamers in India, 74% stated that they prefer casino apps over browser versions, mainly because of the speed and stability offered by platforms like Pin-Up.
Pin Up Slot Collection Explained
The slot segment remains one of the most influential components of the global gaming industry, consistently attracting players with its dynamic mechanics and visual variety. Many users in India gravitate toward titles that blend simplicity with entertainment value, especially when they are supported by strong providers and recognizable themes. This diversity becomes even more appealing within the Pin Up slot category, which is refreshed every month with new releases from leading developers such as Pragmatic Play, NetEnt, Spinomenal, and several others. The steady flow of updates ensures that players never run out of new experiences and always have access to fresh mechanics and features.
A large portion of Indian players prefer medium-volatility slot games because they offer a stable balance between payout frequency and win potential. At the same time, users searching for more intense gameplay can explore high-volatility options that introduce larger but less frequent winning opportunities. For thrill-seekers, jackpot slots introduce the possibility of significant rewards, enhancing the excitement and pushing engagement even further. This range of volatility levels makes the overall collection highly adaptable to the preferences of both beginners and experienced players, strengthening the platform’s appeal and long-term retention.
Types of Slots Available at Pin Up
The platform offers a wide range of slot formats, allowing players to choose games that match their preferred style, pacing, and volatility. This variety is especially important for users who enjoy experimenting with different mechanics, themes, and bonus structures. To help players navigate the catalogue more effectively, the main slot categories available on the platform are grouped into clear segments that highlight the strengths of each format.
- Classic three-reel slots
- Five-reel video slots
- Megaways™ slots
- Branded slots
- Progressive jackpot slots
- Bonus-buy slots
Each type offers unique mechanics and RTP characteristics. For beginners, classic slots provide the easiest entry point. For more experienced players, Megaways™ and progressive jackpots offer advanced features and potentially larger wins.
The slot interface at Pin-Up is designed to be visually appealing while remaining user-friendly. Providers often use retro art combined with modern animation to complement the brand’s vintage theme.
Pin Up Casino Aviator Strategies and Insights
Among all games at Pin-Up, the Pin Up Casino Aviator stands out as the most viral. The game’s simple interface and high adrenaline factor make it a favorite in India, especially among younger players aged 21-30. Aviator is a crash-style game where the multiplier increases until it randomly stops, and the player must cash out before the crash.
The combination of unpredictability and strategy has led to increasing popularity across social media. Indian influencers often highlight Aviator gameplay because of its fast pace and modern feel.
Why Aviator Is Popular Among Indian Players
Many Indian users are drawn to crash-style games because they offer fast, dynamic gameplay that feels different from traditional casino formats. Aviator stands out even more due to its combination of simplicity and high engagement, making it appealing to both beginners and players who prefer strategic decision-making. To understand why the game continues to grow in popularity, it helps to look at the specific features that contribute to its widespread appeal among Indian audiences.
Indian players enjoy Aviator for several reasons:
- Extremely simple rules
- Quick rounds lasting under 10 seconds
- Opportunity to use mathematical strategies
- Social chat where players interact live
- High multipliers with potential large payouts
The transparency of the game and its reliance on probability rather than reels make many players treat it as a mix of entertainment and tactical skill.
A quote from a frequent Indian user illustrates its popularity:
“Aviator gives you both excitement and control — every second matters. “
Pin Up Bonus Casino Promotions and Loyalty Perks
The Pin Up Bonus Casino system is designed to reward both new and returning players. Bonuses vary from welcome packages to weekly reloads, free spins, cashback, and tournament rewards. For Indian users, INR-based promotions make the platform feel more localized and accessible.
Pin-Up also features seasonal promotions tied to major Indian festivals — Diwali, Holi, and New Year. These limited-time events often include free spins, deposit gifts, and leaderboard tournaments.
Types of Bonuses Available at Pin Up
Bonus programs play an essential role in shaping the overall user experience, especially for players who value long-term engagement and added incentives. The platform offers a variety of promotional options that cater to both newcomers and experienced users, ensuring that every stage of gameplay feels rewarding. To help players understand what they can receive while exploring different sections of the platform, the most common bonus types have been grouped into a simple, easy-to-navigate structure.
Below is a list of common bonuses Indian players can expect:
- Welcome bonus package
- Free spins for selected slots
- Weekly cashback
- Daily missions and rewards
- VIP loyalty points
- Tournament prizes
These bonuses motivate users to explore more categories, including slots, live games, and Aviator. Players should always read the wagering requirements to understand how many times the bonus must be played before withdrawal.
Pin-Up ensures transparency by listing all conditions clearly in each offer section, keeping the user experience clean and trustworthy.
FAQ About Pin-Up
What is Pin-Up and how does it work?
Pin-Up is an online entertainment platform offering casino games, slots, Aviator, and live dealer games tailored for Indian users. Players register, deposit funds, choose games, and enjoy various features like bonuses and tournaments.
Is the Pin Up app safe to use in India?
Yes, the Pin Up app uses encrypted technology to protect user data and supports secure payment methods like UPI, PayTM and bank cards, ensuring safe transactions.
Which games are most popular at Pin Up casino?
The most popular games include Pin Up slot titles, live dealer tables, and the highly engaging Pin Up casino aviator, known for its rapid rounds and high multipliers.
How do I claim a Pin Up bonus casino offer?
Players can activate bonuses through the promotions section after registration or deposit. Each bonus includes wagering rules, which must be completed before withdrawal.
Is Aviator based on luck or strategy?
Aviator combines probability with strategic timing. While outcomes are random, strategies such as early cash-outs and controlled bet sizing help manage risk.